#include <sorption_base.hh>
|
| | TYPEDEF_ERR_INFO (EI_ArrayName, std::string) |
| |
| | DECLARE_INPUT_EXCEPTION (ExcSubstanceCountMatch,<< "The size of the input array "<< EI_ArrayName::qval<< " does not match the number of substances.") |
| |
| | SorptionBase (Mesh &init_mesh, Input::Record in_rec) |
| |
| virtual | ~SorptionBase (void) |
| |
| void | initialize () override |
| | Prepares the object to usage. More...
|
| |
| void | zero_time_step () override |
| |
| void | update_solution (void) override |
| | Updates the solution. More...
|
| |
| void | output_data (void) override |
| | Output method. More...
|
| |
| bool | evaluate_time_constraint (FMT_UNUSED double &time_constraint) override |
| |
| | TYPEDEF_ERR_INFO (EI_Substance, std::string) |
| |
| | TYPEDEF_ERR_INFO (EI_Model, std::string) |
| |
| | DECLARE_INPUT_EXCEPTION (ExcUnknownSubstance,<< "Unknown substance name: "<< EI_Substance::qval) |
| |
| | DECLARE_INPUT_EXCEPTION (ExcWrongDescendantModel,<< "Impossible descendant model: "<< EI_Model::qval) |
| |
| | ReactionTerm (Mesh &init_mesh, Input::Record in_rec) |
| | Constructor. More...
|
| |
| | ~ReactionTerm (void) |
| | Destructor. More...
|
| |
| void | choose_next_time (void) override |
| | Disable changes in TimeGovernor by empty method. More...
|
| |
| ReactionTerm & | substances (SubstanceList &substances) |
| | Sets the names of substances considered in transport. More...
|
| |
| ReactionTerm & | output_stream (std::shared_ptr< OutputTime > ostream) |
| | Sets the output stream which is given from transport class. More...
|
| |
| virtual bool | evaluate_time_constraint (double &time_constraint)=0 |
| | Computes a constraint for time step. More...
|
| |
| ReactionTerm & | concentration_fields (FieldFEScalarVec &conc_mobile) |
| |
| | EquationBase () |
| |
| | EquationBase (Mesh &mesh, const Input::Record in_rec) |
| |
| virtual | ~EquationBase () |
| |
| virtual void | set_time_upper_constraint (double dt, std::string message) |
| |
| virtual void | set_time_lower_constraint (double dt, std::string message) |
| |
| TimeGovernor & | time () |
| |
| virtual void | set_time_governor (TimeGovernor &time) |
| |
| double | planned_time () |
| |
| double | solved_time () |
| |
| Mesh & | mesh () |
| |
| std::shared_ptr< Balance > | balance () const |
| |
| TimeMark::Type | mark_type () |
| |
| FieldSet & | data () |
| |
Definition at line 59 of file sorption_base.hh.
Constructor with parameter for initialization of a new declared class member
Definition at line 124 of file sorption_base.cc.
| SorptionBase::~SorptionBase |
( |
void |
| ) |
|
|
virtual |
| SorptionBase::SorptionBase |
( |
| ) |
|
|
protected |
This method disables to use constructor without parameters.
| void SorptionBase::clear_max_conc |
( |
| ) |
|
|
protected |
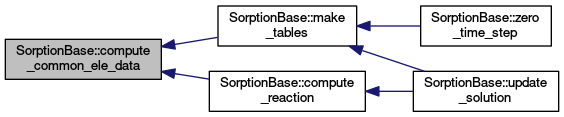
| virtual void SorptionBase::compute_common_ele_data |
( |
const ElementAccessor< 3 > & |
elem | ) |
|
|
protectedpure virtual |
| SorptionBase::DECLARE_INPUT_EXCEPTION |
( |
ExcSubstanceCountMatch |
, |
|
|
<< "The size of the input array "<< EI_ArrayName::qval<< " does not match the number of substances." |
|
|
) |
| |
| bool SorptionBase::evaluate_time_constraint |
( |
FMT_UNUSED double & |
time_constraint | ) |
|
|
inlineoverride |
| const Record & SorptionBase::get_input_type |
( |
| ) |
|
|
static |
Static variable for new input data types input
Definition at line 54 of file sorption_base.cc.
| void SorptionBase::initialize |
( |
| ) |
|
|
overridevirtual |
Prepares the object to usage.
Allocating memory, reading input, initialization of fields.
Reimplemented from EquationBase.
Definition at line 165 of file sorption_base.cc.
| void SorptionBase::initialize_fields |
( |
| ) |
|
|
protected |
| void SorptionBase::initialize_from_input |
( |
| ) |
|
|
protected |
Initializes private members of sorption from the input record.
Definition at line 253 of file sorption_base.cc.
| void SorptionBase::initialize_substance_ids |
( |
| ) |
|
|
protected |
Reads names of substances from input and creates indexing to global vector of substance.
Also creates the local vector of molar masses.
Definition at line 207 of file sorption_base.cc.
| void SorptionBase::isotherm_reinit |
( |
unsigned int |
i_subst, |
|
|
const ElementAccessor< 3 > & |
elm |
|
) |
| |
|
protected |
Reinitializes the isotherm.
On data change the isotherm is recomputed, possibly new interpolation table is made. NOTE: Be sure to update common element data (porosity, rock density etc.) by compute_common_ele_data(), before calling reinitialization!
Definition at line 399 of file sorption_base.cc.
| static Input::Type::Instance SorptionBase::make_output_type |
( |
const string & |
equation_name, |
|
|
const string & |
output_field_name, |
|
|
const string & |
output_field_desc |
|
) |
| |
|
inlinestatic |
| void SorptionBase::make_reactions |
( |
| ) |
|
|
protected |
Initializes possible following reactions from input record. It should be called after setting mesh, time_governor, distribution and concentration_matrix if there are some setting methods for reactions called (they are not at the moment, so it could be part of init_from_input).
Definition at line 138 of file sorption_base.cc.
| void SorptionBase::make_tables |
( |
void |
| ) |
|
|
protected |
| void SorptionBase::output_data |
( |
void |
| ) |
|
|
overridevirtual |
Output method.
Some reaction models have their own data to output (sorption, dual porosity)
- this is where it must be reimplemented. On the other hand, some do not have (linear reaction, pade approximant)
- that is why it is not pure virtual.
Reimplemented from ReactionTerm.
Definition at line 576 of file sorption_base.cc.
| void SorptionBase::set_initial_condition |
( |
| ) |
|
|
protected |
Reads and sets initial condition for concentration in solid.
Definition at line 363 of file sorption_base.cc.
| SorptionBase::TYPEDEF_ERR_INFO |
( |
EI_ArrayName |
, |
|
|
std::string |
|
|
) |
| |
| void SorptionBase::update_max_conc |
( |
| ) |
|
|
protected |
Computes maximal aqueous concentration at the current step.
Definition at line 448 of file sorption_base.cc.
| void SorptionBase::update_solution |
( |
void |
| ) |
|
|
overridevirtual |
Updates the solution.
Goes through local distribution of elements and calls compute_reaction.
Reimplemented from EquationBase.
Definition at line 379 of file sorption_base.cc.
| void SorptionBase::zero_time_step |
( |
| ) |
|
|
overridevirtual |
Does first computation after initialization process. The time is set and initial condition is set and output.
Reimplemented from EquationBase.
Definition at line 337 of file sorption_base.cc.
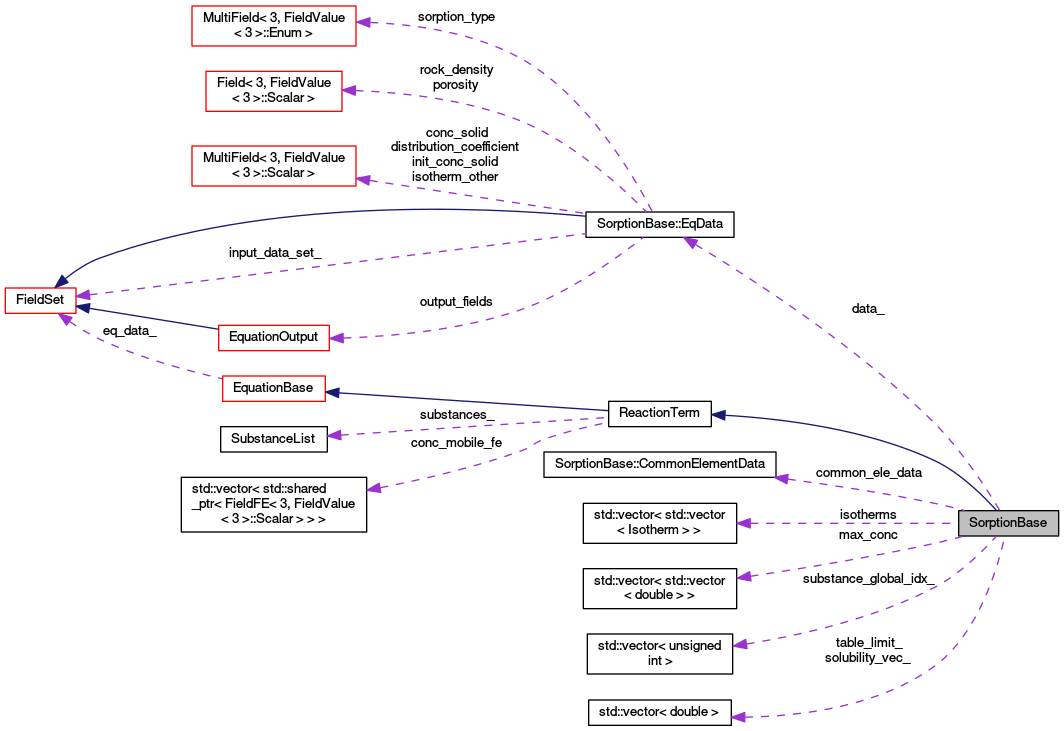
Pointer to equation data. The object is constructed in descendants.
Definition at line 193 of file sorption_base.hh.
Three dimensional array contains intersections between isotherms and mass balance lines. It describes behaviour of sorbents in mobile pores of various rock matrix enviroments. Up to |nr_of_region x nr_of_substances x n_points| doubles. Because of equidistant step lenght in cocidered system of coordinates, just function values are stored.
Definition at line 223 of file sorption_base.hh.
Maximum concentration per region. It is used for optimization of interpolation table.
Definition at line 216 of file sorption_base.hh.
| unsigned int SorptionBase::n_interpolation_steps_ |
|
protected |
Temporary nr_of_points can be computed using step_length. Should be |nr_of_region x nr_of_substances| matrix later.
Definition at line 198 of file sorption_base.hh.
| unsigned int SorptionBase::n_substances_ |
|
protected |
Critical concentrations of species dissolved in water.
Definition at line 207 of file sorption_base.hh.
| double SorptionBase::solvent_density_ |
|
protected |
Density of the solvent. TODO: Could be done region dependent, easily.
Definition at line 203 of file sorption_base.hh.
| std::vector<unsigned int> SorptionBase::substance_global_idx_ |
|
protected |
Concentration table limits of species dissolved in water.
Definition at line 211 of file sorption_base.hh.
The documentation for this class was generated from the following files:
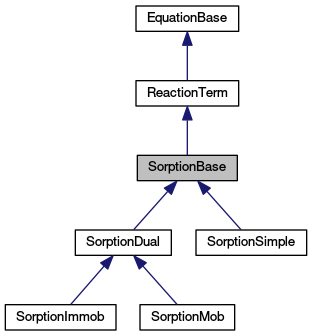
 Public Member Functions inherited from ReactionTerm
Public Member Functions inherited from ReactionTerm Public Member Functions inherited from EquationBase
Public Member Functions inherited from EquationBase Static Public Member Functions inherited from ReactionTerm
Static Public Member Functions inherited from ReactionTerm Static Public Member Functions inherited from EquationBase
Static Public Member Functions inherited from EquationBase Protected Attributes inherited from ReactionTerm
Protected Attributes inherited from ReactionTerm Protected Attributes inherited from EquationBase
Protected Attributes inherited from EquationBase Public Types inherited from ReactionTerm
Public Types inherited from ReactionTerm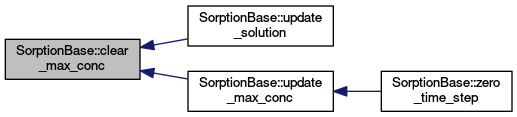







 1.8.11
1.8.11