Base class for linear reactions and decay chain. More...
#include <first_order_reaction_base.hh>
Public Member Functions | |
| FirstOrderReactionBase (Mesh &init_mesh, Input::Record in_rec) | |
| Constructor. More... | |
| ~FirstOrderReactionBase (void) | |
| Destructor. More... | |
| void | initialize () override |
| Prepares the object to usage. More... | |
| void | zero_time_step () override |
| Moves the model to zero time. More... | |
| void | update_solution (void) override |
| Updates the solution. More... | |
| bool | evaluate_time_constraint (double &time_constraint) override |
| Computes a constraint for time step. More... | |
 Public Member Functions inherited from ReactionTerm Public Member Functions inherited from ReactionTerm | |
| TYPEDEF_ERR_INFO (EI_Substance, std::string) | |
| TYPEDEF_ERR_INFO (EI_Model, std::string) | |
| DECLARE_INPUT_EXCEPTION (ExcUnknownSubstance,<< "Unknown substance name: "<< EI_Substance::qval) | |
| DECLARE_INPUT_EXCEPTION (ExcWrongDescendantModel,<< "Impossible descendant model: "<< EI_Model::qval) | |
| ReactionTerm (Mesh &init_mesh, Input::Record in_rec) | |
| Constructor. More... | |
| ~ReactionTerm (void) | |
| Destructor. More... | |
| virtual void | output_data (void) override |
| Output method. More... | |
| void | choose_next_time (void) override |
| Disable changes in TimeGovernor by empty method. More... | |
| ReactionTerm & | substances (SubstanceList &substances) |
| Sets the names of substances considered in transport. More... | |
| ReactionTerm & | output_stream (std::shared_ptr< OutputTime > ostream) |
| Sets the output stream which is given from transport class. More... | |
| ReactionTerm & | concentration_matrix (double **concentration, Distribution *conc_distr, int *el_4_loc, int *row_4_el) |
 Public Member Functions inherited from EquationBase Public Member Functions inherited from EquationBase | |
| EquationBase () | |
| EquationBase (Mesh &mesh, const Input::Record in_rec) | |
| virtual | ~EquationBase () |
| virtual void | set_time_upper_constraint (double dt, std::string message) |
| virtual void | set_time_lower_constraint (double dt, std::string message) |
| TimeGovernor & | time () |
| virtual void | set_time_governor (TimeGovernor &time) |
| double | planned_time () |
| double | solved_time () |
| Mesh & | mesh () |
| TimeMark::Type | mark_type () |
| FieldSet & | data () |
| virtual void | get_solution_vector (double *&vector, unsigned int &size) |
| virtual void | get_parallel_solution_vector (Vec &vector) |
Protected Member Functions | |
| virtual void | assemble_ode_matrix (void)=0 |
| Assembles the matrix of the ODEs. More... | |
| virtual double ** | compute_reaction (double **concentrations, int loc_el) override |
| Computes the reaction on a specified element. More... | |
| virtual void | initialize_from_input ()=0 |
| Initializes private members of sorption from the input record. More... | |
| unsigned int | find_subst_name (const std::string &name) |
 Protected Member Functions inherited from ReactionTerm Protected Member Functions inherited from ReactionTerm | |
| virtual void | output_vector_gather (void) |
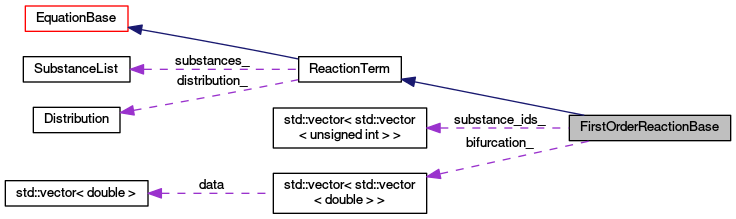
Protected Attributes | |
| std::vector< std::vector< unsigned int > > | substance_ids_ |
| std::vector< std::vector< double > > | bifurcation_ |
| unsigned int | n_substances_ |
| Number of all transported substances. It is the dimension of the reaction matrix. More... | |
| arma::mat | reaction_matrix_ |
| Reaction matrix. More... | |
| arma::vec | prev_conc_ |
| Column vector storing previous concetrations on an element. More... | |
| arma::mat | molar_matrix_ |
| Diagonal matrix with molar masses of substances. More... | |
| arma::mat | molar_mat_inverse_ |
Inverse of molar_matrix_. More... | |
| std::shared_ptr< PadeApproximant > | linear_ode_solver_ |
 Protected Attributes inherited from ReactionTerm Protected Attributes inherited from ReactionTerm | |
| double ** | concentration_matrix_ |
| int * | el_4_loc_ |
| Indices of elements belonging to local dofs. More... | |
| int * | row_4_el_ |
| Indices of rows belonging to elements. More... | |
| Distribution * | distribution_ |
| Pointer to reference to distribution of elements between processors. More... | |
| SubstanceList | substances_ |
| Names belonging to substances. More... | |
| std::shared_ptr< OutputTime > | output_stream_ |
| Pointer to a transport output stream. More... | |
 Protected Attributes inherited from EquationBase Protected Attributes inherited from EquationBase | |
| bool | equation_empty_ |
| flag is true if only default constructor was called More... | |
| Mesh * | mesh_ |
| TimeGovernor * | time_ |
| Input::Record | input_record_ |
| FieldSet * | eq_data_ |
| std::shared_ptr< Balance > | balance_ |
| object for calculation and writing the mass balance to file. More... | |
Additional Inherited Members | |
 Static Public Member Functions inherited from ReactionTerm Static Public Member Functions inherited from ReactionTerm | |
| static Input::Type::Abstract & | it_abstract_term () |
| static Input::Type::Abstract & | it_abstract_mobile_term () |
| static Input::Type::Abstract & | it_abstract_immobile_term () |
| static Input::Type::Abstract & | it_abstract_reaction () |
Detailed Description
Base class for linear reactions and decay chain.
The class implements common interface for linear reactions and decay chains. One step of the linear reaction or the decay is represented as a product of a reaction matrix and a vector of concentrations of transported substances on a single element.
It uses armadillo to compute the reaction matrix which then multiplies to concetration vector. This class also resolves the choice of the numerical method which is used to compute the reaction matrix.
Definition at line 40 of file first_order_reaction_base.hh.
Constructor & Destructor Documentation
| FirstOrderReactionBase::FirstOrderReactionBase | ( | Mesh & | init_mesh, |
| Input::Record | in_rec | ||
| ) |
Constructor.
Definition at line 40 of file first_order_reaction_base.cc.
| FirstOrderReactionBase::~FirstOrderReactionBase | ( | void | ) |
Destructor.
Definition at line 47 of file first_order_reaction_base.cc.
Member Function Documentation
|
protectedpure virtual |
Assembles the matrix of the ODEs.
We solve the system of  equations
equations
![\[ \frac{\textrm{d} c_i}{\textrm{d}t}=-\sum\limits_{j=1}^N \frac{M_i}{M_j} \lambda_{i} b_j c_j, \qquad \textrm i=1,\ldots,N \]](form_21.png)
where  are the molar masses of the parent substances and products,
are the molar masses of the parent substances and products,  are the reaction rate constants (in case of decays converted from half_lives) and
are the reaction rate constants (in case of decays converted from half_lives) and  are the branching ratios. The constant coefficients
are the branching ratios. The constant coefficients  are the elements of the system matrix.
are the elements of the system matrix.
It is pure virtual and must be implemented in descendants.
Implemented in RadioactiveDecay, and FirstOrderReaction.

|
overrideprotectedvirtual |
Computes the reaction on a specified element.
Implements ReactionTerm.
Definition at line 91 of file first_order_reaction_base.cc.

|
overridevirtual |
Computes a constraint for time step.
Implements ReactionTerm.
Definition at line 137 of file first_order_reaction_base.cc.
|
protected |
Help function to create mapping of substance indices. Finds a position of a string in specified array.
Definition at line 127 of file first_order_reaction_base.cc.
|
overridevirtual |
Prepares the object to usage.
Allocating memory, reading input, initialization of fields.
Reimplemented from EquationBase.
Definition at line 51 of file first_order_reaction_base.cc.
|
protectedpure virtual |
Initializes private members of sorption from the input record.
Implemented in FirstOrderReaction, and RadioactiveDecay.

|
overridevirtual |
Updates the solution.
Goes through local distribution of elements and calls compute_reaction.
Reimplemented from EquationBase.
Definition at line 110 of file first_order_reaction_base.cc.
|
overridevirtual |
Moves the model to zero time.
The assembly of the system matrix is called here.
Reimplemented from EquationBase.
Definition at line 77 of file first_order_reaction_base.cc.
Member Data Documentation
|
protected |
Two dimensional array contains mass percentage of every single decay bifurcation on every single row.
Definition at line 103 of file first_order_reaction_base.hh.
|
protected |
Definition at line 114 of file first_order_reaction_base.hh.
|
protected |
Inverse of molar_matrix_.
Definition at line 112 of file first_order_reaction_base.hh.
|
protected |
Diagonal matrix with molar masses of substances.
Definition at line 111 of file first_order_reaction_base.hh.
|
protected |
Number of all transported substances. It is the dimension of the reaction matrix.
Definition at line 106 of file first_order_reaction_base.hh.
|
protected |
Column vector storing previous concetrations on an element.
Definition at line 109 of file first_order_reaction_base.hh.
|
protected |
Reaction matrix.
Definition at line 108 of file first_order_reaction_base.hh.
|
protected |
Sequence of integers describing an order of isotopes. substance_ids_[reactant][local_product_idx] = global_substance_idx
Definition at line 98 of file first_order_reaction_base.hh.
The documentation for this class was generated from the following files:
- /opt/flow123d/flow123d/src/reaction/first_order_reaction_base.hh
- /opt/flow123d/flow123d/src/reaction/first_order_reaction_base.cc
 1.8.11
1.8.11